
|
Introduction |
|
|
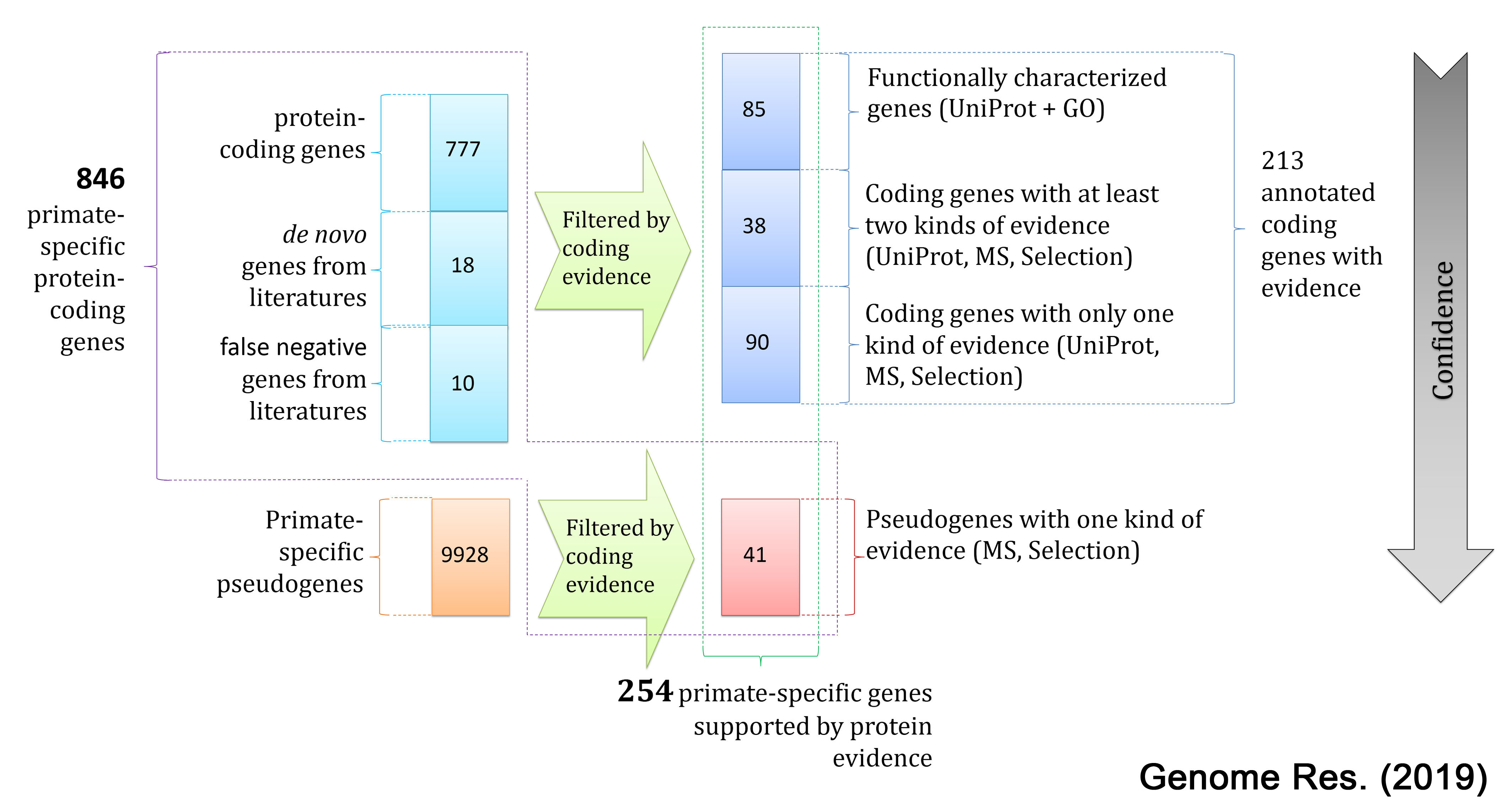
New genes have frequently formed and spread to fixation in a wide variety of organisms, constituting abundant sets of lineage-specific genes. Recent reports show counter-intuitive evidence that new genes quickly assume critical roles in the brain and developmental pathways of primates and Drosophila, indicating that the genetics of these essential structures and processes can evolve rapidly. More generally, these findings suggest that genomes are continually evolving in both sequence and content, eroding the conservation endowed by common ancestry. Despite increasing recognition of the importance of new genes, these genes are still seriously under-characterized in functional studies and that new gene annotation is inconsistent in current practice. In order to alleviate this situation, we are developing GenTree asa central portal to catalog lineage-specific genes in various species. As a first step, we present the age information for protein-coding genes in human together with the inferred origination mechanism. Later on, we will expand to other species such as mouse and fruitfly and import multiple lines of evidence on whether the currently annotated gene model is correct. As of now, GenTree loaded human data onto the database, which is based on Ensembl v73 (Sep. 2013). In addition, fly age data based on Ensembl v78 (Dec. 2014) is also available for the bulk download. |
|
|
|
|
Citation
|
|
|
Human Data: Shao Y., et al. (2019). GenTree, an integrated resource for analyzing the evolution and function of primate-specific coding genes. Genome Res 29: 682-696. [PubMed] Fly Data: Zhang Y. E., et al. (2010). Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res 20(11): 1526-1533. [PubMed] |
| Last Modified at April 26, 2019 Copyright@ 2013-2026 by the ZhangLab colleagues, CAS, Any Comments and suggestions mail to:zhanglabioz@gmail.com |